I’ve tried to run it, but I’m getting error
ERROR: ' ' does not exist in current working directory ('/Users/shijianhui').
I don’t really get what I’m doing wrong 
I’ve tried to run it, but I’m getting error
ERROR: ' ' does not exist in current working directory ('/Users/shijianhui').
I don’t really get what I’m doing wrong 
Here’s a small version of my notes that I used for testing this code; maybe anyone who is interested in trying it out can make sure that they get results from this one first to make sure that there are no issues with the install (or my instructions). Then, if still keen, I can try and help debug any issues that occur (post tbx file here or dm me?) in other files that I likely did not think of when only testing against my notes.
sample-tbx-file.tbx (103.7 KB)
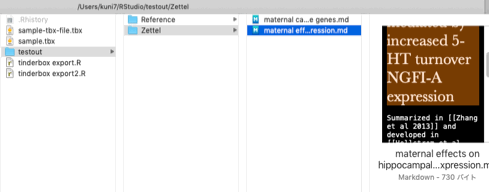
After running the code above on this sample file the output should look like this:
testout/
testout/Reference
testout/Reference/Hellstrom et al 2012.md
testout/Reference/Zhang et al 2013.md
testout/Zettel
testout/Zettel/maternal care regulates a host of NGFI-A sensitive genes.md
testout/Zettel/maternal effects on hippocampal glucocorticoid receptors mediated by increased 5-HT turnover NGFI-A expression.md
Of course, I should reiterate as @satikusala nicely summarized in his documentation/videos of Tinderbox export and @mwra has documented in depth elsewhere , my xml parser is not likely ever going to compete with proper exports. It was me struggling with export plus, mostly, trying to put the no-data-lockin principle to the test (which it passed with flying colours - I now know I can get very useful data out of tinderbox even if I somehow lose access to tinderbox). Still, happy to make that script more usable if anyone thinks they would benefit.
Super interesting exercise. Maybe you can come to our Saturday meet up and show us. So far, I’ve not been able to get it to work, even with the test file you’ve shared. Clearly, my setup is wrong.
Dear jpl,
Thanks a lot sharing your sample-tbx-file.tbx.
I try your R.script with RStudio from home.
I am afraid that lack of facility with something .
As it turned this [can not read xml.png].
My circumstance : Mac OS 10.14.6 Mojave R ver4.0.5 RStudio ver.1.4.1106
Tinderbox 8.6.2
Sincerely yours, WAKAMATSU
Dear jpl,
R can not find function [read_xml].
This message should put these old processes to work in better ways.
How can I get this behaviour { find function [read_xml]}?
Thx and regards, WAKAMATSU
Dear jpl,
I got it.
Two additions were made,
and the result was a successful compilation.
No.01 : At the front of R.script. , I add 3 loading lines.
library(xml2)
library(tidyverse)
library(stringi)
No.02 : And similar additions in L85.
L 85 : litnotes <- xml2::read_xml(“sample.tbx”)
I add xml2:: ahead of read_xml
Now I can sleep in peace.
Thx and regards, WAKAMATSU
@jpl Very nice! Here the output doesn’t go to md files but to the console.
So some more code needed to write to .md files, right?
I notice you have only internal links in the sample TBX file, which all get converted to [[wikilinks]]. How would one handle a document with a mixture of internal and web links, where one would want [] for internal links and anchor for web links? The only way I could figure out how to address that in the first script in this thread (which addresses a similar problem as your script here but assumes a working copy of the app is available) was to add a @ prefix to internal links. Having looked at the xml, do you see a better way to distinguish between the two kinds of links?
@satikusala I was reminded in this return to R after several years that the Run button in RStudio runs the selected lines only. So you might try making sure you have selected the whole script before hitting Run. I’m guessing you may have missed one or more of those three crucial library() lines at the start. (Thereafter you can just select the lines you want to rerun (as in my screenshot).
Ah, that makes sense.
I now am getting an error on tidyverse. Any idea on what I can do to install it properly. I’m new to RStudio.
What worked for me was to type (not copy-paste) these into the terminal, being careful to have straight quotes…
Here tidyverse took a long time (many minutes) to install, with multiple messages appearing in the console, as it has many parts. The other two were quicker.
If no success, then doing something like what is described here might help.
Hi all - it seems like mixed success at best for my somewhat terrible code/instructions mix. I’ll try and find some time in the next few days to create a proper R packages with a little interface to make it easier to use. Sorry about the clumsy first attempt.
I suspect that can be solved by running
install.packages("colorspace")
and then trying again? (Keep quote substitutions in my example in mind - any hints on how to avoid those?)
The final function, write_to_md, should output to file.
And sure, I think it’d be possible to add a proper anchor for web links; for the moment (since it fit my use case) I only modified the internal links and otherwise left the code entirely alone. I also ignore the rtfd part, so if the links are embedded in there I’d first have to figure out how to deal with rtf …
Out curiosity, @jpl, if you were to do it in python, which packages would you use?
BTW, I tried on a dummy file and it worked perfectly  On my actual Tbx file it didn’t though. I think it is because the links don’t always match the name of the note being linked.
On my actual Tbx file it didn’t though. I think it is because the links don’t always match the name of the note being linked.
Ya! I got it working, sort-of. I can get the output now, but it goes to the console not to the files on the drive. Thanks.
It didn’t create the files? For me it both outputs what you see and creates the files as well (i.e. ‘testout/Reference/Zhang et al 2013.md’ should now exist).
Nope. Just output to the console. This is so super cool! Thanks for doing this. And, thank ou all for your patience as I’ve learned R. Prett cool.
Bizarre. Can you (in a terminal) run ‘ls -alF’ on the directory you are testing it in? Is it at least creating the base directories that it is supposed to?
The directories are created on my drive.
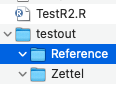
But when I run what you ask I get:
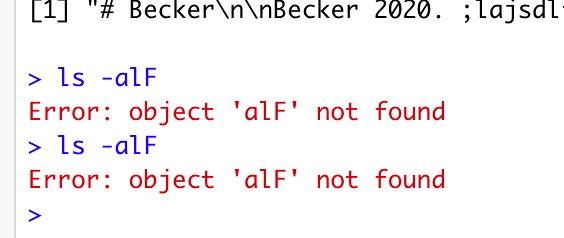
I’m stumped - I have no idea why it wouldn’t create the actual files for you. Maybe when I get around to cleaning up the code it’ll become easier to figure out, or otherwise feel free to send me the file you used and the code (in case you made any modifications), but for the moment I suspect I’ll remain stumped.
(And I meant it as a shell command in the Mac Terminal, not the RStudio terminal, but the screenshot was enough).