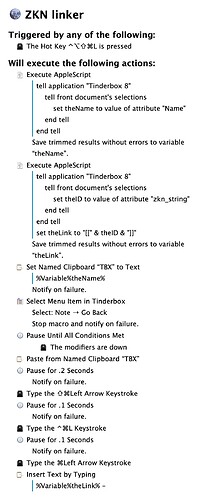
It turns out I’m too dumb to fully grok the tinderbox export mechanisms. But this prompted me to follow through on the promise of tinderbox not locking you in via having parsable xml files. Turns out the file format is quite beautiful and parsable! I thus created some code that takes all my tinderbox notes and spits out directories of markdown files, replete with wikilinks. I did this to play with noteplan3 a bit, but the files are equally readable (and links work) in obsidian and devonthink. For the sake of anyone who wants to do something similar I’ve added my code below.
A few notes:
- this only takes the plain text part of each note, ignores any rtf formatting
- it assumes that the link text and target id name are the same thing (which was true for me due to creating all links via zip links; would be quite easy to move away from that assumption).
- assumes that interesting items can be selected via their prototypes.
The code is in R - would be just as easy in python or ruby or whatever, but I went with what I’ve used more recently. Needs the tidyverse, xml2, and stringi packages.
Really not sure this is useful to anyone, but just in case, here it is:
library(xml2)
library(tidyverse)
library(stringi)
### function definitions
add_wikilinks <- function(ID, text, links_df) {
# find relevant links
links <- links_df %>% filter(sstart>0 & sourceid==ID) %>% arrange(sstart)
# return existing text if no links were found
if (nrow(links)==0) {
return(text)
}
# create substring: takes the link location and surrounds it by [[]]
# this assumes that the source name and link text are the same.
# then returns the modified text string
wikified <- paste0('[[',
stri_sub(text, from=links$sstart+1, length=links$slen),
']]')
return(stri_sub_replace_all(text,
from=links$sstart+1,
length=links$slen,
value=wikified))
}
tbx_links <- function(tbx_xml) {
# grab all link entries
links <- xml_find_all(tbx_xml, ".//link")
# convert link endtries to data frame, keeping link name sourceid, destid, and start and length
links_df <- data.frame(
name=links %>% xml_attr("name"),
sourceid=links %>% xml_attr("sourceid"),
destid=links %>% xml_attr("destid"),
sstart=links %>% xml_attr("sstart") %>% as.numeric(),
slen=links %>% xml_attr("slen") %>% as.numeric()
)
return(links_df)
}
tbx_items <- function(tbx_xml, links_df) {
# grab all item entries
items <- xml_find_all(tbx_xml, ".//item")
# convert to dataframe
items_df <- data.frame(
# get item ID
ID=items %>% xml_attr("ID"),
# get item prototype
proto=items %>% xml_attr("proto"),
# find first Name attribute nested below this item
Name=items %>% xml_find_first(".//attribute[@name='Name']") %>% xml_text(),
# find first text entry nested below this item
text=items %>% xml_find_first(".//text") %>% xml_text()
) %>% # now add the wikilinks and header text
mutate(wikilinks=map2_chr(ID, text, add_wikilinks, links_df=links_df),
wikitext=paste0('# ', Name, '\n\n', wikilinks))
return(items_df)
}
write_to_md <- function(items_df, outpath, prototypes) {
# check output directories, create if necessary
dirs <- paste(outpath, prototypes, sep="/")
for (d in dirs) {
if (!dir.exists(d)) {
dir.create(d, recursive = TRUE)
}
}
# take the items
items_df %>%
# keep just the one with the prespecified prototypes
filter(proto %in% prototypes) %>%
# and write each to file
split(.$ID) %>%
map(~ write_file(.x$wikitext, file=paste0(outpath, "/", .x$proto, "/", .x$Name, ".md"), append = FALSE))
}
### Run it for my data
# start by reading my tbx file
litnotes <- read_xml("~/Dropbox/litnotes.tbx")
# create a dataframe of all the links
links_df <- tbx_links(litnotes)
# create a dataframe of all the items; this includes wikilinkifying
items_df <- tbx_items(litnotes, links_df)
# write out those items with Zettel and Reference prototypes
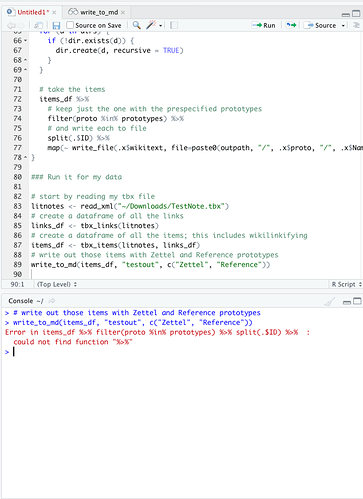
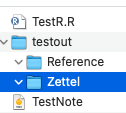
write_to_md(items_df, "testout", c("Zettel", "Reference"))